Hi,
a new tutorial on the analysis pan-genomes using
GET_HOMOLOGUES and GET_HOMOLOGUES-EST is now available. After a short introduction, where
the main concepts are illustrated, the remaining sections cover the
installation and typical operations required to analyze and annotate
genomes and transcriptomes from a pan-genome perspective, in which
individuals or species contribute genetic material to a pool.
The examples include both bacterial sequences in GenBank format and plant transcripts. This tutorial has been created for a two-day workshop to be held at BIOS (Manizales, Colombia) next week, with title "From genomes to pangenomes: understanding variation among individuals and species":
The tutorial can be found at: http://digital.csic.es/handle/10261/146411
Code, sample datasets and documentation are available at:
https://github.com/eead-csic-compbio/get_homologues
Suggestions and error reports are welcome,
Bruno
Ideas y código para problemas de genómica de plantas, biología computacional y estructural
9 de marzo de 2017
13 de febrero de 2017
Actualización de Algoritmos3D
Hola,
sirva esta entrada para publicar la versión actualizada del material sobre "Algoritmos en Bioinformática estructural", que podéis encontrar en:
http://eead-csic-compbio.github.io/bioinformatica_estructural
Este material lo usamos anualmente en la licenciatura en ciencias genómicas de la UNAM en el campus de Cuernavaca, México.
Un saludo,
Bruno
sirva esta entrada para publicar la versión actualizada del material sobre "Algoritmos en Bioinformática estructural", que podéis encontrar en:
http://eead-csic-compbio.github.io/bioinformatica_estructural
Este material lo usamos anualmente en la licenciatura en ciencias genómicas de la UNAM en el campus de Cuernavaca, México.
 |
| Predicción de estructuras protéicas a partir de sus patrones de mutaciones correlacionadas en secuencias homólogas, tomada de http://science.sciencemag.org/content/355/6322/248.full. Ests tipo de protocolos se presentan en la sección http://eead-csic-compbio.github.io/bioinformatica_estructural/node35.html. |
Un saludo,
Bruno
5 de enero de 2017
HS-BLASTN as replacement of MEGABLAST
Hi,
this is hopefully the last of a series of posts where I explored software choices that might replace BLAST+ programs in some scenarios. Today I'll write about HS-BLASTN, a parallel nucleotide local aligner reported to accelerate the default BLASTN algorithm (megablast), while producing the same results:
Note that megablast is a fast choice for intra-species comparisons and typically retrieves sequence matches with nucleotide identities greater than 70% (see Figure 3 in http://eead-csic-compbio.github.io/get_homologues/manual-est).
We will benchmark HS-BLASTN (version hs-blastn-0.0.5+) using the same Hordeum vulgare and Arabidopsis thaliana sequences used in a previous post.
It can be seen that indexing a FASTA file with 67K sequences is about 16x slower with HS-BLASTN than with standard NCBI mableblastdb, and produces much larger index files (629Mb vs 28Mb). Now let's review search performance:
It can be seen that HS-BLASTN is ~4x faster using a single thread. About the same speedup is obtained when 20 threads are used and BLASTN is parallelized with help from an external script (_split_blast.pl). There are small nuisances, such as the fact that no soft-masking is available, or the ocasional non-stable output order of hits with same score, but it seems worth it nevertheless,
have a good weekend,
Bruno
this is hopefully the last of a series of posts where I explored software choices that might replace BLAST+ programs in some scenarios. Today I'll write about HS-BLASTN, a parallel nucleotide local aligner reported to accelerate the default BLASTN algorithm (megablast), while producing the same results:
 | |
| HS-BLASTN algorithm overview, taken from http://nar.oxfordjournals.org/content/early/2015/08/06/nar.gkv784.long |
We will benchmark HS-BLASTN (version hs-blastn-0.0.5+) using the same Hordeum vulgare and Arabidopsis thaliana sequences used in a previous post.
$ hs-blastn index bur-0.fasta #[IndexBuilder] Time elapsed: 89.002952 secs. #-rw-rw-r-- 1 contrera contrera 442M Jan 5 09:59 bur-0.fasta.bwt #-rw-rw-r-- 1 contrera contrera 13M Jan 5 09:58 bur-0.fasta.header #-rw-rw-r-- 1 contrera contrera 116M Jan 5 09:59 bur-0.fasta.sa #-rw-rw-r-- 1 contrera contrera 58M Jan 5 09:58 bur-0.fasta.sequence $ ncbi-blast-2.2.30+/bin/makeblastdb -in bur-0.fasta -dbtype nucl #Adding sequences from FASTA; added 67259 sequences in 5.51543 seconds. #-rw-rw-r-- 1 contrera contrera 12M Jan 5 10:06 bur-0.fasta.nhr #-rw-rw-r-- 1 contrera contrera 789K Jan 5 10:06 bur-0.fasta.nin #-rw-rw-r-- 1 contrera contrera 15M Jan 5 10:06 bur-0.fasta.nsq
It can be seen that indexing a FASTA file with 67K sequences is about 16x slower with HS-BLASTN than with standard NCBI mableblastdb, and produces much larger index files (629Mb vs 28Mb). Now let's review search performance:
hs-blastn align -query SBCC073_fLF.fasta \ -db bur-0.fasta -evalue 0.00001 -outfmt 6 -max_target_seqs 5 \ -out SBCC073_fLF.bur-0.hsblastn #[HS-BLASTN] done. Elpased time: 9.0055 secs. hs-blastn align -query SBCC073_fLF.fasta \ -db bur-0.fasta -evalue 0.00001 -outfmt 6 -max_target_seqs 5 \ -out SBCC073_fLF.bur-0.hsblastn -num_threads 20 #[HS-BLASTN] done. Elpased time: 1.6599 secs. time ncbi-blast-2.2.30+/bin/blastn -task megablast -query SBCC073_fLF.fasta \ -db bur-0.fasta -evalue 0.00001 -soft_masking true -outfmt 6 -max_target_seqs 5 \ -out SBCC073_fLF.bur-0.blastn #real 0m33.943s perl _split_blast.pl 20 2000 ncbi-blast-2.2.30+/bin/blastn \ -task megablast -query SBCC073_fLF.fasta -db bur-0.fasta -evalue 0.00001 -soft_masking true \ -outfmt 6 -max_target_seqs 5 -out SBCC073_fLF.bur-0.blastn # runtime: 7 wallclock secs ( 2.81 usr 0.50 sys + 60.47 cusr 7.28 csys = 71.06 CPU)
It can be seen that HS-BLASTN is ~4x faster using a single thread. About the same speedup is obtained when 20 threads are used and BLASTN is parallelized with help from an external script (_split_blast.pl). There are small nuisances, such as the fact that no soft-masking is available, or the ocasional non-stable output order of hits with same score, but it seems worth it nevertheless,
have a good weekend,
Bruno
19 de diciembre de 2016
DIAMOND as alternative to BLASTP
Hi,
in a recent post I discussed the performance of DIAMOND when aligning nucleotide sequences against a peptide database. However, as this tool can also do protein similarity searches, I wanted to test that too. Here I summarize those results.
We used all protein sequences encoded in ecotype Bur-0 of Arabidopsis thaliana (n=53,027) and the proteome of Mycobacterium tuberculosis strain H37Rv (n=3,906). We scanned these against Swissprot, downloaded from ftp.uniprot.org and formatted as follows:
As earlier, we used v0.8.25, obtained from https://github.com/bbuchfink/diamond.
The actual sequence similarity searches were performed as follows:
The take home message is that DIAMOND "more sensitive" is 20x to 100x faster than BLASTP in these tests, with roughly 15% less sensitive overall, which is reduced to 5-9% when more remote homologues matter.
Bruno
PS I post another test done by David A Wilkinson, from Massey University, New Zealand:
"I ran get homologues in conjunction with blastp, diamond in "standard mode" and diamond in "more-precise mode" for a small dataset.The dataset includes 20 genomes from the genus Leptospira - importantly, they are not all from the same "species", and for some pairwise comparisons will have relatively distant genomes and gene identities...
in a recent post I discussed the performance of DIAMOND when aligning nucleotide sequences against a peptide database. However, as this tool can also do protein similarity searches, I wanted to test that too. Here I summarize those results.
$ makeblastdb -in uniprot_sprot.fasta -dbtype prot $ diamond makedb --in uniprot_sprot.fasta --db uniprot_sprot.fasta
As earlier, we used v0.8.25, obtained from https://github.com/bbuchfink/diamond.
The actual sequence similarity searches were performed as follows:
$ diamond blastp -p 30 -d swissprot -q bur-0.faa -o bur-0.diamond.tsv \ --max-target-seqs 1000 --evalue 0.00001 \ --outfmt 6 qseqid sseqid pident length mismatch gapopen qstart qend \ sstart send evalue bitscore qcovhsp sseq $ diamond blastp -p 30 -d swissprot -q bur-0.faa -o bur-0.diamond.sensitive.tsv \ --sensitive ... $ diamond blastp -p 30 -d swissprot -q bur-0.faa -o bur-0.diamond.more.tsv \ --more-sensitive ... $ time ncbi-blast-2.2.30+/bin/blastp -num_threads 30 -db swissprot \ -query bur-0.faa -out bur-0.blastp.tsv -max_target_seqs 1000 -evalue 0.00001 -outfmt '6 std qcovhsp sseq'
# calling _split_blast.pl as used in get_homologues
$ get_homologues-est/_split_blast.pl 30 250 ncbi-blast-2.2.30+/bin/blastp \ -db swissprot -query bur-0.faa -out bur-0.split.blastp.tsv \ -max_target_seqs 1000 -evalue 0.00001 -outfmt \'6 std qcovhsp sseq\'Those commands were then run replacing bur-0.faa with Mtuberculosis_H37Rv.faa and using 8 CPU cores instead of 30. The obtained alignments were compared with a script (available upon request). Wall-clock time and matched hits are computed in relation to BLASTP (control). Alignment comparisons were performed with all BLASTP hits (red) and the subset of hits with identity >= 50% (blue).
The take home message is that DIAMOND "more sensitive" is 20x to 100x faster than BLASTP in these tests, with roughly 15% less sensitive overall, which is reduced to 5-9% when more remote homologues matter.
search_strategy time(s) matched =length longer shorter matched =length longer shorter
control (NCBI BLASTP) 16440 [ all hits ] [ IDENTITY:50% ]
bur-0.diamond 55 0.402 0.804 0.109 0.088 0.848 0.916 0.063 0.021
bur-0.diamond.sensitive 192 0.774 0.794 0.116 0.090 0.906 0.916 0.062 0.022
bur-0.diamond.more 263 0.832 0.797 0.115 0.088 0.910 0.916 0.062 0.022
bur-0.split.blastp 6915 1.000
control (NCBI BLASTP) 2657 [ all hits ] [ IDENTITY:50% ]
Mtub.diamond 34 0.483 0.853 0.075 0.072 0.930 0.932 0.051 0.017
Mtub.diamond.sensitive 124 0.832 0.823 0.091 0.087 0.957 0.931 0.052 0.017
Mtub.diamond.more 141 0.853 0.821 0.092 0.086 0.958 0.930 0.052 0.017
Mtub.split.blastp 2334 1.000
Bruno
PS I post another test done by David A Wilkinson, from Massey University, New Zealand:
"I ran get homologues in conjunction with blastp, diamond in "standard mode" and diamond in "more-precise mode" for a small dataset.The dataset includes 20 genomes from the genus Leptospira - importantly, they are not all from the same "species", and for some pairwise comparisons will have relatively distant genomes and gene identities...
I used the ORTHOMCL algorithm in get-homologues for clustering. My numbers are in total agreement with published literature.Here are the comparative results:
#clusters
|
#core
|
#softcore
|
#shell
|
#cloud
|
|
BLASTP
|
10398
|
1523
|
2197
|
2269
|
5932
|
DIAMOND_STANDARD
|
10829
|
1421
|
2146
|
2328
|
6355
|
DIAMOND_MORE_PRECISE
|
10410
|
1535
|
2215
|
2269
|
5926
|
%clusters
|
%core
|
%softcore
|
%shell
|
%cloud
|
|
BLASTP
|
100%
|
100%
|
100%
|
100%
|
100%
|
DIAMOND_STANDARD
|
104%
|
93%
|
98%
|
103%
|
107%
|
DIAMOND_MORE_PRECISE
|
100%
|
101%
|
101%
|
100%
|
100%
|
"
15 de diciembre de 2016
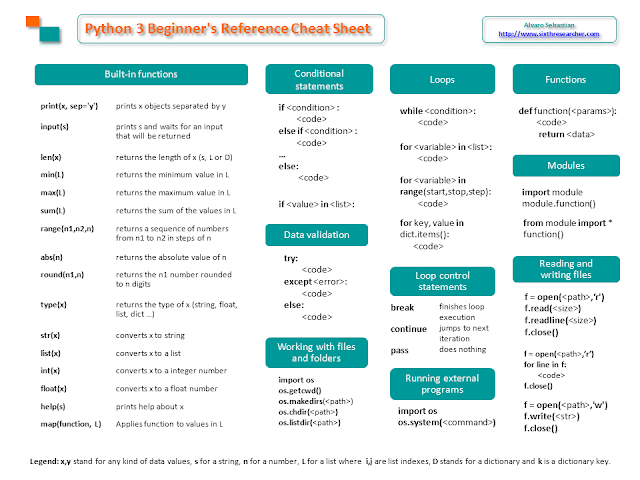
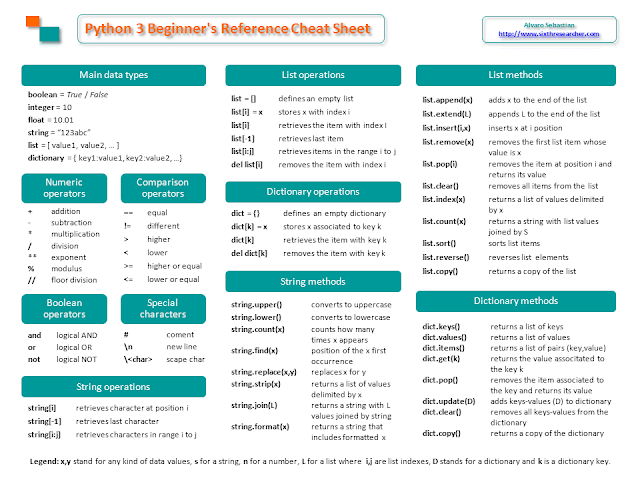
Chuleta de Python 3 para principiantes
Hoy quiero compartir con vosotros una chuleta de Python 3 que he creado para mis estudiantes, he intentado recopilar en dos caras de DIN-A4 los tipos de datos, operadores, métodos, funciones y otros contenidos útiles que uno necesita tener a mano cuando comienza a programar en Python 3 (incluso meses más tarde).
Puedes descargarte la chuleta en formato PDF de mi blog en inglés o usarla online:
Puedes descargarte la chuleta en formato PDF de mi blog en inglés o usarla online:
Suscribirse a:
Entradas (Atom)