Elaine Harrison empieza explicando el programa científico de ELIXIR, articulado alrededor de tres temas de la nueva capa/tier científica:
1. Patrick Aloy (ES) introduce el tema "Biología celular y molecular" e insiste en ir más allá de los datos para poder reconstruir el "full analytic journey" de cada estudio.
2. Robert Waterhouse (CH) introduce el tema que nos toca más cerca "Biodiversity, food security, & pathogens (BFSP)"
3. Serena Scollen (Hub) presenta "Human data and translational research" y el ciclo completo de los datos genómicos humanos
Taller "Single-cell galaxy user journey" with Wendi Bacon.
https://galaxyproject.org/community/sig/singlecell
https://www.biostars.org/p/471274
No data standard yet, closest is https://anndata.readthedocs.io (really an HDF5 derivative python friendly, R users still use dataframes).
https://usegalaxy.eu workflows can be created graphically, or by stacking up and exporting the operations you carried out on your data.
Workflows are ultimately text files; easy to convert workflows galaxy -> snakemake / nextflow by exporting to bash, difficult the other way around. Can be stored at https://workflowhub.eu
https://github.com/galaxyproject/idc -> genomic references for Galaxy
https://biostar.galaxyproject.org/p/11944/index.html
"Defer dataset" allows using public URL as input, data only downloaded when executing in particular galaxy node, only results stored in main; saves disk quota.
Taller "Interop_Mini-Symposium_All_Hands_2024"
RDA = Research Data Alliance
ebi.ac.uk/metabolights , still lack accepted standards for data deposition.
Wei Kheng Teh talks about metadata heterogeneity of single-cell omics data
https://isa-tools.org/ -> https://simplifier.net/guide/isa-to-fhir?version=current
https://www.researchobject.org/ro-crate/ -> lightweight approach to packaging research data with their metadata, see https://doi.org/10.5281/zenodo.5146227, can be stored in Zenodo or GitHub, Workflow -> to get work done / Dataflow -> to publish and share data.
ELIXIR-CZ are working on writing material / templates for DMPs, there's a gap there, on bridging high level interoperability aims and actual detailed protocols.
Interoperability barriers: traditional rules, increasing data complexity, we still need a minimal standard for acrossfields data integration, heterogeneity of data quality.
Taller "Paving the way towards the effective use of generative AI for ELIXIR - Agenda"
Michael Hu, PI and Director of Bioinformatics at West Virginia University habla de "Bioinformatics with ChatGPT"
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1011511
https://arxiv.org/abs/2403.15274
Renat Shigapov, Data Scientist at University of Mannheim talks about "Reviewing (meta)data and evaluate their FAIRness using ChatGPT+", pero su charla requiere una licencia de chatGTP Plus. La idea central es que pare que un conjunto de datos sea FAIR entonces deben poder encontrarlo tanto otras personas como software y que GPT puede ayudar en esa tarea, siempre y cuando se incluyan conexiones a fuentes externas para comprobar las URLs que devuelve y evitar alucionaciones. Sugiere que ELIXIR debería usar herramientas como https://github.com/UB-Mannheim/FAIR-GPT
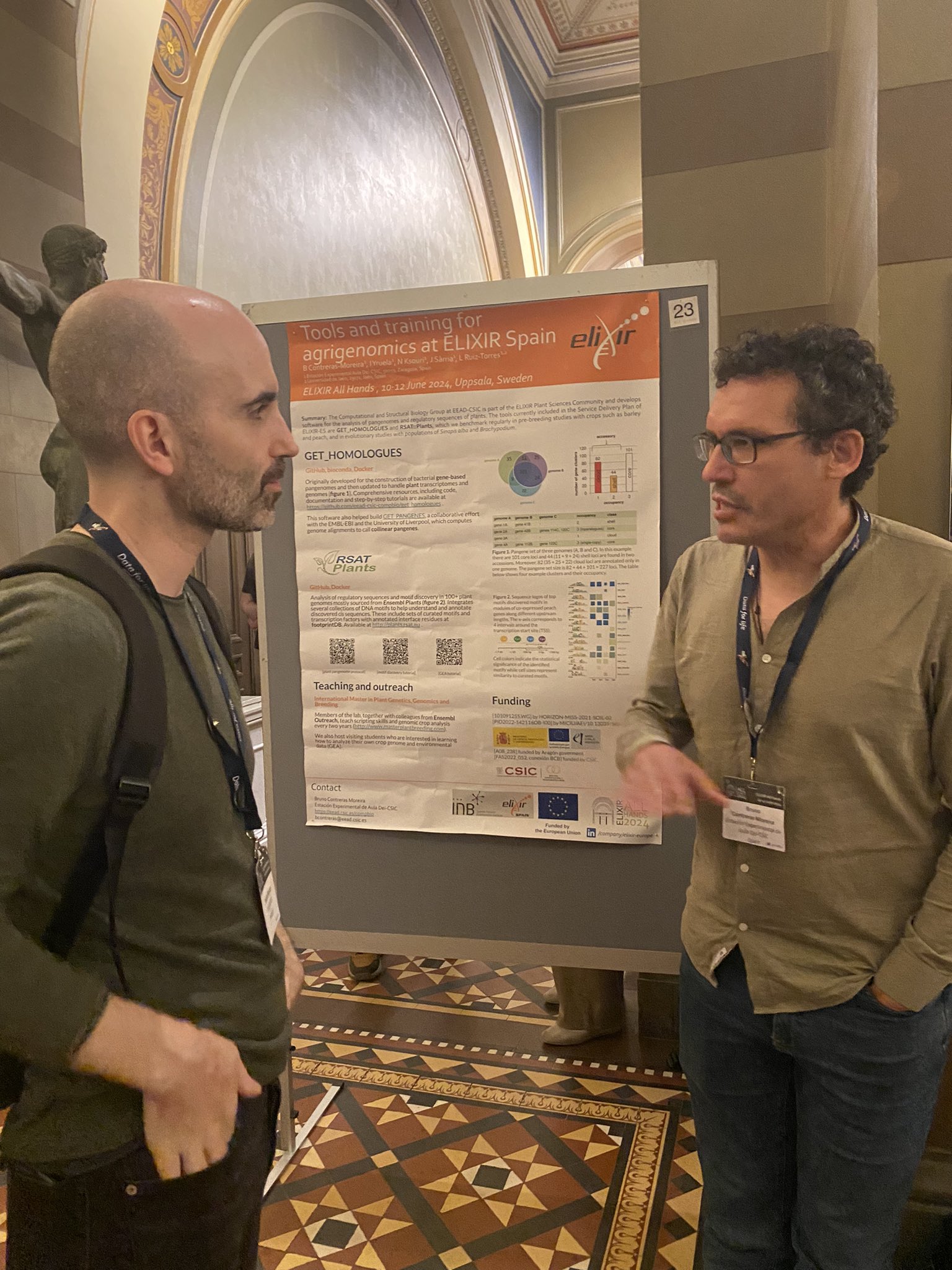 |
| Poster disponible en https://doi.org/10.7490/f1000research.1119714.1, foto de Ana Conesa https://x.com/anaconesa/status/1800828390607610058 |
PD 12062024 day III
ELIXIR technical tier, 5 Plataforms
Ejemplos de proy EU que se convirtieron en servcios: RDMkit, workflowhub
M Jetten presenta RDM community
Elixir software registry: bio.tools > openebench > biocontainers > galaxy
https://github.com/research-software-ecosystem/content
TeSS training support system https://tess.elixir-europe.org
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007854
LS Login
No hay comentarios:
Publicar un comentario