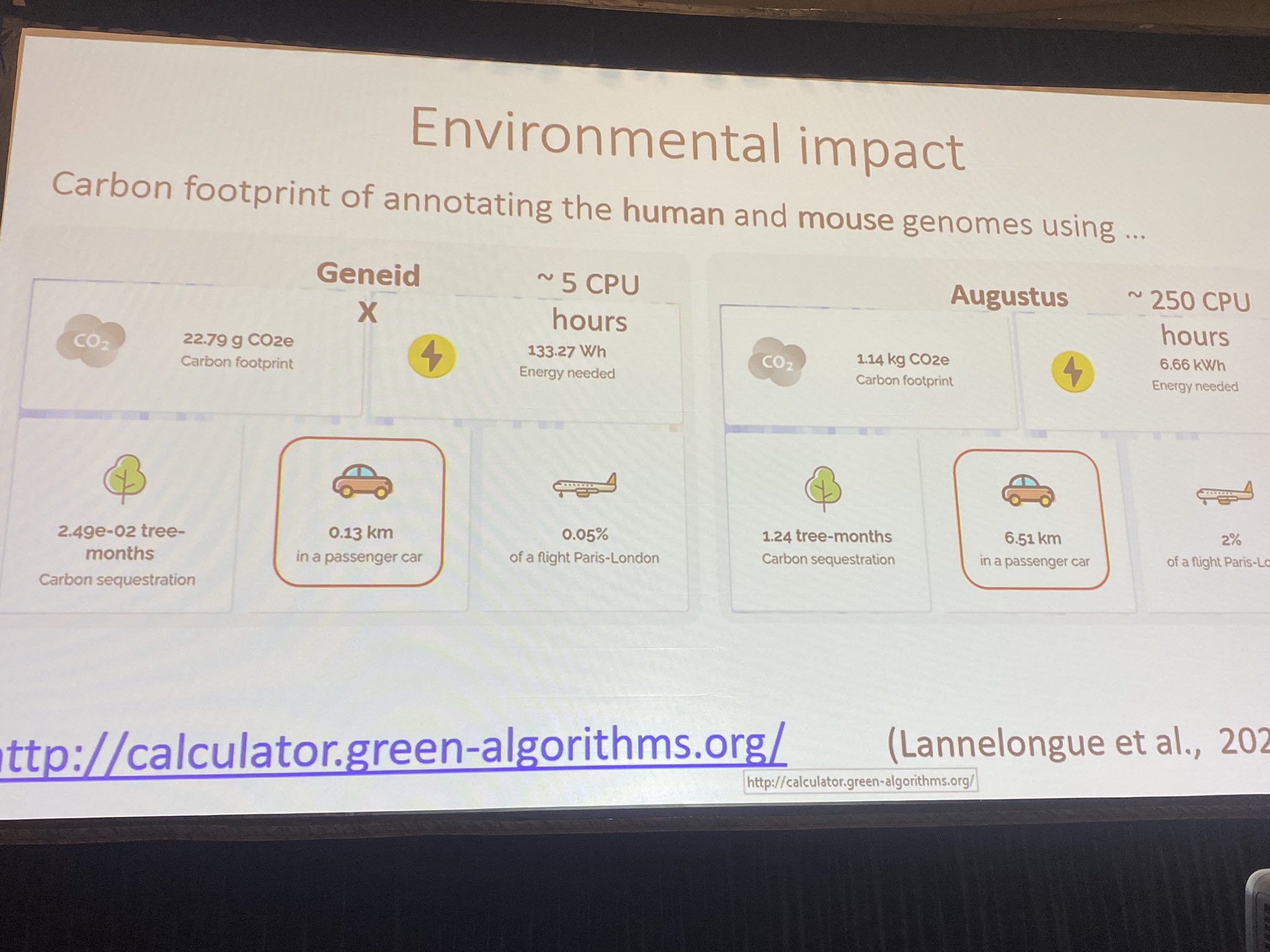
Wednesday 18012023
Oliver Ryder, San Diego Zoo
Institute for Conservation Research. He talks about conservation genomics. For
instance, he explains how the genomes of extinct species can be used to
estimate genetic load and effective population size. On extant species, genome
sequences can be used to estimate heterozygosity for instance (Californian
condor > Andean condor). He then moves to technological possibilities to
rescue/clone[nearly-]extinct species if we are able to crioperserve DNA and
tissues. San Diego’s zoo has its own FrozenZoo for this.
Thomas Blein, Institute of
Plant Sciences Paris-Saclay. MARS lncRNA regulates the coexpression of a gene
cluster, recruits a TF to a promoter, controls a chromatin loop and coordinates
epigenetically the expression of neighbor genes, in response to ABA: https://pubmed.ncbi.nlm.nih.gov/35150931 . You can read there that “The
enrichment of co-regulated lncRNAs in clustered metabolic genes in Arabidopsis
suggests that the acquisition of novel non-coding transcriptional units may
constitute an additional regulatory layer driving the evolution of biosynthetic
pathways”.
Peijian Cao, China Tobacco
Gene Research Center. Starts by presenting lncRNAs that encode small peptides (https://academic.oup.com/bib/article/20/5/1853/5047384, http://14.139.61.8/PlantPepDB/index.php). In his experiments they track the
expression of lncRNAs in response to herbivores. They find that different
lncRNAs coexpress with genes in the biosynthetic pathway to that produces JA.
They also have evidence of encoded small peptides being produced upon binding
to ribosomes. These peptides are as stable as common proteins.
Hikmet Budak, Montana BioAg
Inc. Presents work on lncRNAs on different wheats to try and control pest insects.
Josephine Herbst, VIB-UGent
Center for Plant Systems Biology.