Hi, last week the AlphaFold3 paper came out in the journal Nature among concerns about the code and data not being available, some by sources as close as referee #3 of the manuscript (read more here and here).
I couldn't find the time to test it with some plant transcription factors (TF) we have worked with over the years. Then I found this thread and could wait no more.
So I took barley VRN1, which according to our own footprintDB database binds to the consensus CCarAAAwGG, as determined by ChIPseq by Deng et al:
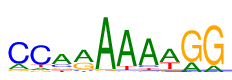 |
| Sequence logo for VRN1 |
As this is a SRF-type/MADS box TF, known to bind as dimers (see for instance complex 1hbx_AB), I logged in at the Alphafold3 beta server at https://golgi.sandbox.google.com/about and pasted the following input. Note that I padded the DNA duplex with ACGT at both sides and replaced degenerate positions with adenines (having several targets on the same DNA duplex might confuse AF3, see here):
Once the job finished I downloaded and uncompressed the 2.9Mb results file and computed logos and DNA motifs for all 5 produced models using the Docker container at https://hub.docker.com/r/eeadcsiccompbio/dnaprot:
# convert CIF to PDB files conda install -c conda-forge gemmi gemmi convert fold_2024_05_10_16_53_model_0.cif vrn1_0.pdb ... gemmi convert fold_2024_05_10_16_53_model_4.cif vrn1_4.pdb # actually get DNA motifs and sequence logos docker run --rm -v "$PWD:$PWD" -w "$PWD" -u $UID:$GROUPS \
eeadcsiccompbio/dnaprot dnaprot.pl -P ./VRN1_0 -i vrn1_0.pdb
...
docker run --rm -v "$PWD:$PWD" -w "$PWD" -u $UID:$GROUPS \ eeadcsiccompbio/dnaprot dnaprot.pl -P ./VRN1_4 -i vrn1_4.pdb
I got the 5 sequence DNA motifs, one for each AF3 model:
# IC=8.461 IC/col=0.769
A | 0 0 0 24 57 96 57 24 0 0 53
C | 1 96 96 24 12 0 12 24 0 0 12
G | 6 0 0 24 12 0 12 24 96 96 12
T | 89 0 0 24 15 0 15 24 0 0 19
# IC=5.996 IC/col=0.545
A | 4 0 4 24 24 24 84 24 0 16 48
C | 4 96 92 24 24 24 4 24 4 20 16
G | 0 0 0 24 24 24 4 24 92 56 12
T | 88 0 0 24 24 24 4 24 0 4 20
Which correspond to the following sequence logos:
In summary, it seems that the AF3 models fed with a cognate DNA sequence can be used to produce reasonable DNA motifs (compared to the consensus CCarAAAwGG). However, note that the nucleotides immediately before and after the consensus seem to be also moderately conserved in the motifs, and in this case they come from the padding sequences. While this indicates that padding can affect the obtained logos, the logo published by Deng et al did include one of those bases.
PS See also this thread for membrane proteins: https://twitter.com/jankosinski/status/1789062090205921768

https://www.science.org/content/article/limits-access-deepmind-s-new-protein-program-trigger-backlash
ResponderEliminarhttps://www.linkedin.com/feed/update/urn:li:activity:7263948687779762176/
ResponderEliminarhttps://www.nature.com/articles/s41586-025-09422-z
ResponderEliminar